SPARE-SEA
Environmental Spread and Persistence of Antibiotic Resistances in aquatic Systems Exposed to oyster Aquaculture

Partners
Partners
CNRS, Ifremer, Université de Montpellier, Université de Perpignan (CNRS)
National Research Council (CNR)
University of Genova (UNIGE)
Institut De Recerca I Tecnología Agroalimentaria (IRTA)
Abstract
Abstract
Aquaculture has been identified as a gateway for antibiotic resistance (AR) spread, but little is known of AR in the oyster aquaculture environment. The biggest oyster aquaculture industry cultivates the Pacific oyster Crassostrea gigas, which is cultured in marine coastal areas that are often contaminated by AR determinants (antibiotics, resistance genes, and resistant bacteria) and other pollutants.
Moreover, antibiotics are used in hatcheries, and since oysters accumulate bacteria, the consumption of raw oysters can be a vector for AR into human microbiomes. Also, AR transmission to other species threatens the safety of coastal marine systems, the sustainability of shellfish farming and human health.
By combining human, animal and environmental health, SPARE-SEA implements a One Health approach to identify environmental drivers and pathways of AR spread within and between environmental compartments including known and emerging pathogens.
By investigating the cumulative effects of human use of coastal ecosystems along multiple gradients (e.g oyster farming intensity and agrochemical pollutant run-off) on the enrichment of AR in the oyster bio-reactor and its subsequent transfer routes within anthroposized coastal environments, we will link objectives of all three JPIs involved.
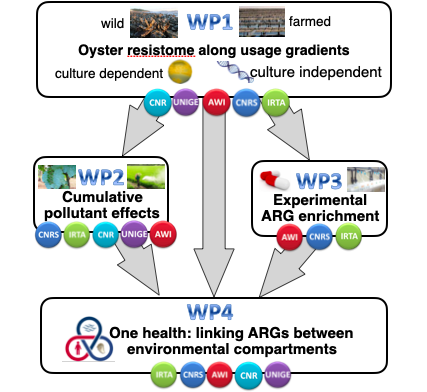
WP structure
WP structure
WP number | Led by |
| WP1 | UNIGE |
| WP2 | CNRS |
| WP3 | AWI |
| WP4 | IRTA |
Expected research results
Expected research results
- Improved knowledge on Antibiotic-resistant bacteria in oysters
- Inventory of zoonotic ARB