MAPMAR
Marine Plasmids Driving the Spread of Antibiotic Resistances

Partners
Partners
National Center for Mariculture (NCM), Israel Oceanographic and Limnological Research
Karlsruhe Institute of Technology (KIT), Institute for Biological Interfaces
Abstract
Abstract
Antibiotic resistance genes (ARGs) are one of the most challenging contaminants of emerging concern (CECs). Instead of being directly produced by human activity, ARGs emerge as consequence of antibiotic use in clinical settings, and residual antibiotic contamination. ARGs spread through horizontal gene transfer and conjugative plasmids, because their ability to cross inter-species barriers are key in this process.
Recent findings revealed the existence of marine plasmids (MAPS) of global distribution and broad host range. These MAPS can transmit ARGs across oceanic distances and may reintroduce them to human food chains via marine products. They are, however, different to classical plasmids from clinical settings.
MAPMAR uses metagenomics, data science and single-cell sequencing to obtain a catalog of most prevalent and transmissible MAPs. By testing methods to block their transmission, MAPMAR explores strategies to curtail the risk of oceans acting as highways for ARG propagation.
WP structure
WP structure
WP number | Led by |
| WP1 | UC |
| WP2 | KIT |
| WP3 | UC |
| WP4 | NCM |
| WP5 | UC |
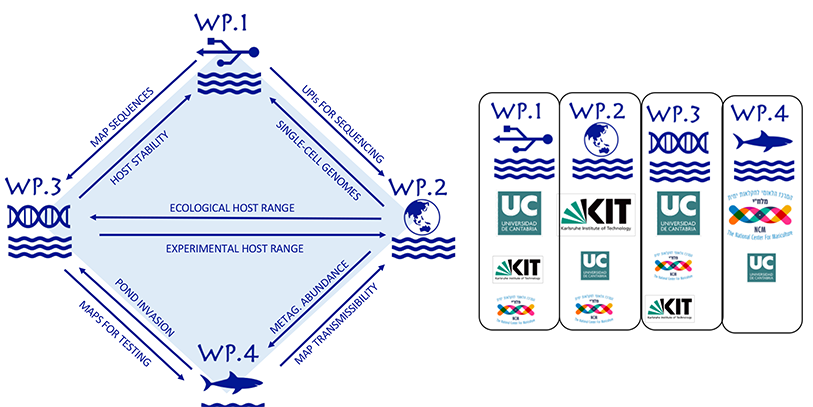
Expected research results
Expected research results
- Identification of plasmids involved in the transmission of antibiotic resistances in the marine environment;
- Assesment of the risk of antibiotic resistance propagation through marine plasmids to the environment;
- Assesment of the risk of antibiotic resistance propagation through marine plasmids to humans through the food chain;
- Assesment of the efficiency of conjugation inhibitors in preventing antibiotic resistance propagation in mariculture activities.